Generating Reference Bitstring using HIPPOS-genref¶
Generating Reference Bitstring with Backbone (Default Setting)¶
Reference bitstring is an essential requirement for similarity coefficient (eg. Tanimoto or McConnaughey coefficient) calculation, which is the common method for comparing the interaction fingerprinting of a test compound and a reference (native ligand) interactions on certain protein.
To generate Reference bitstring, first of all, you need to open the command prompt and enter the
examples\01-na_reference folder. This example uses the Neuraminidase enzyme
for two reasons, first, it is one of the enzymes used in DUD-E (Directory of Useful
Decoy Enhanced) therefore you could use it to measure the effect of interaction fingerprinting
on the enrichment factor. Second, it can demonstrate all of the seven interaction types in
interaction fingerprinting.
As you can see there are three folders and two txt configuration files. Each folder represents
a crystal structure of Neuraminidase, and contain the original PDB file and the
split component (protein, ligand, and water) generated with
SPORES. PDB file alone
can not be used as the reference, it has to be in mol2 or pdbqt to ensure that the
atom typing, charge assignment, and protonation is identical to the docking environment
(whether for PLANTS or VINA). So the PDB files here only act as
the source if you want to use PDBQT files as the reference instead. 1b9s,
1b9t, and 1b9v are the PDB ID of the same Neuraminidase, where each of them
bound to different ligand (FDI, RAI, and RA2 respectively). Therefore
the protein name and ligand name should use protein.mol2 and the corresponding ligand
file name as you can see in genref-config.txt.
# first residue is 77
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
proteins 1b9s/protein.mol2 1b9t/protein.mol2 1b9v/protein.mol2
ligands 1b9s/ligand_FDI468_0.mol2 1b9t/ligand_RAI468_0.mol2 1b9v/ligand_RA2468_0.mol2
outfile ref-results.txt
The first line is merely the commented line, everything started with # sign will
be ignored by hippos and hippos-genref. If you open the mol2 protein file with a text editor
you will see that the first residue is Glutamate with residue number 77. However, when
the file is read by Openbabel it will count as residue number 1.
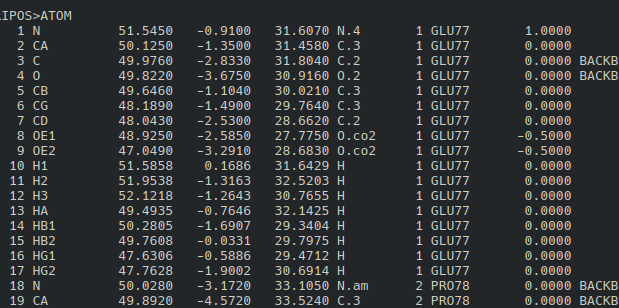
This is where things started to get tricky, because we have to supply both residue_name
and residue_number properly, or else it will not work as to how we want it to be.
residue_name can be acquired easily by converting your protein into mol2 format, while
the corresponding residue_number must be retrieved from the column before residue name
(eg. residue name ARG116 and GLU117 correspond to residue number 40 and 41 respectively):
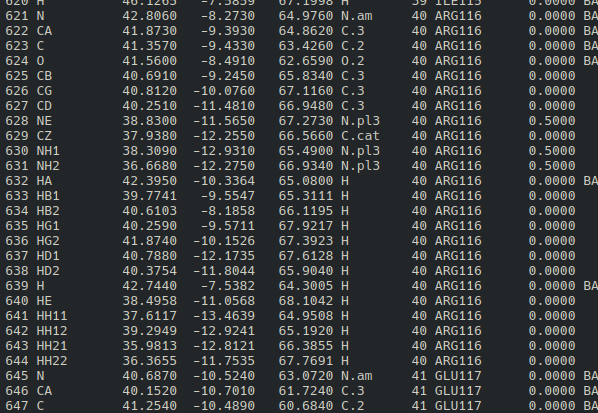
The residue_name and residue_number in genref-config above are retrieved by
visualizing any residue within 5 angstroms from the native ligand using VMD (you can use
any other molecule visualization tool), regardless of how important the residue in
enzyme inhibition.
The next lines are proteins and ligands, notice that there are 3 protein molecules
and 3 ligand molecules which means that there are 3 protein-ligand pairs as references. Where
the first protein will be matched with first ligand and so on. However in most cases, one
protein-ligand pair is enough, this example uses 3 protein-ligand pairs as a demonstration
of multiple references.
The last line is the output file name, it is optional so when not defined the output file will be genref-results.txt.
After we understand the input file and the configuration file, hippos-genref could be run with the following command:
hippos-genref genref-config.txt
After hippos-genref finished file ref-results.txt will be generated.
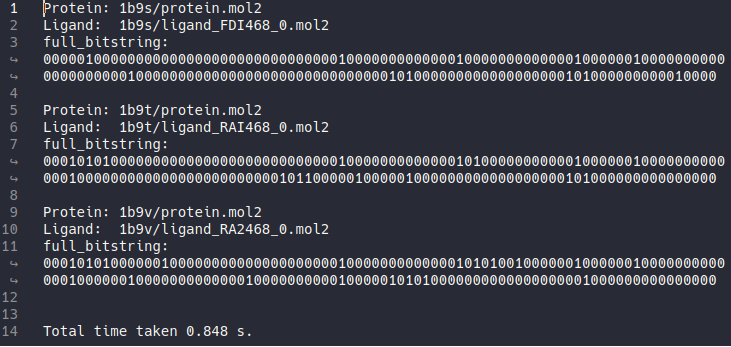
Inside ref-results.txt we can see that there are 3 results from 3 protein-ligand pairs. Each result consisted of protein-ligand pair name and interaction bitstring where each residue represented by 7 bit of interactions from both the side-chain and the backbone.
Generating Reference Bitstring without Backbone¶
Sometimes we would like to omit the interaction between ligand and the backbone protein.
In that case, we should change the output_mode to full_nobb by adding
output_mode full_nobb to our hippos-genref config file as appear in genref-config-nobb.txt
# first residue is 77
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
proteins 1b9s/protein.mol2 1b9t/protein.mol2 1b9v/protein.mol2
ligands 1b9s/ligand_FDI468_0.mol2 1b9t/ligand_RAI468_0.mol2 1b9v/ligand_RA2468_0.mol2
output_mode full_nobb
outfile ref-results-nobb.txt
Now run hippos-genref again with the following command:
hippos-genref genref-config-nobb.txt
After hippos-genref finished file ref-results-nobb.txt will be generated.
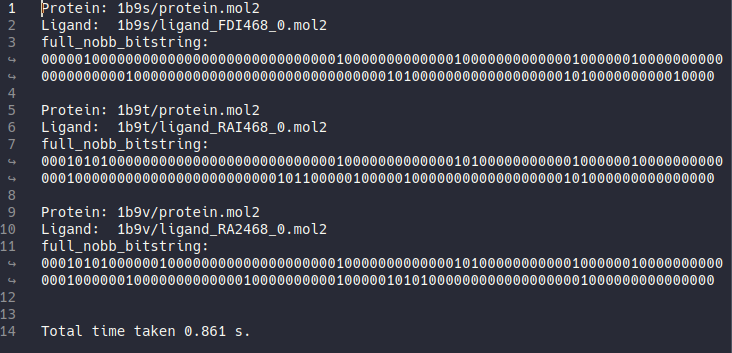
Just like in the default setting, it will generate 3 results. And although they appear the same as before, this time the bitstrings are generated without taking backbone atoms into account.
Generating Simplified Reference Bitstring¶
It is also possible to calculate simplified interaction between ligand and the backbone protein.
In that case, we should change the output_mode to simplified by adding
output_mode simplified to our hippos-genref config file as appear in genref-config-simplified.txt
# first residue is 77
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
proteins 1b9s/protein.mol2 1b9t/protein.mol2 1b9v/protein.mol2
ligands 1b9s/ligand_FDI468_0.mol2 1b9t/ligand_RAI468_0.mol2 1b9v/ligand_RA2468_0.mol2
output_mode simplified
outfile ref-results-simplified.txt
Now run hippos-genref again with the following command:
hippos-genref genref-config-simplified.txt
After hippos-genref finished file ref-results-simplified.txt will be generated.
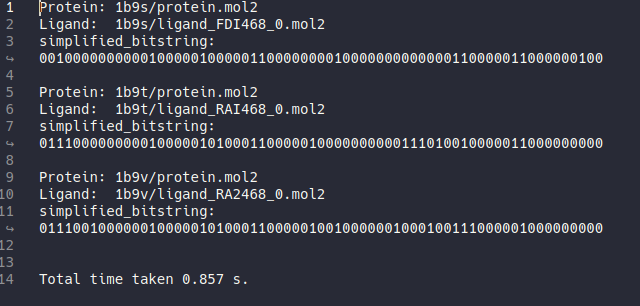
Just like in the default setting, it will generate 3 results. And although they appear the same as before, this time the bitstrings are simplified.