Getting Started (HIPPOS on PLANTS)¶
There are two ways to use HIPPOS, the first one is to use HIPPOS without any reference, and the second one is to use HIPPOS with reference and calculate the similarity coefficient against the reference.
Generating Protein-Ligand Interaction Bitstring without reference¶
First, enter the examples folder and check the configuration example for this
method, config-plants-na-notc.txt
docking_method plants # plants or vina
docking_conf 02-na_plants/plants-003.conf
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
full_outfile plants_notc_ifp.csv
logfile plants_notc.log
The configuration here is pretty much self-explaining. docking_method here is plants,
which correspond to the docking result we would like to analyse. Then the docking_conf
is the configuration file used for docking, HIPPOS require this file to find the details
about docking input and output from PLANTS.
For residue_name and residue_number check the
explanation for the configuration on generating reference with Hippos-genref
for more details.
Next, you can run HIPPOS by entering the following command:
hippos config-plants-na-notc.txt
After the calculation finished HIPPOS will generate 2 files, hippos.log and plants_notc_ifp.csv. plants_notc.log contain the information about ligand name, number of poses, and the running time. If output_mode set to simplified or combo there will be a table for bit position for each residue (useful for deciphering the simplified bitstring). The plants_notc_ifp.csv file will contain the ligand name, pose number, energy from docking result, and the interaction bitstring as can be seen below:
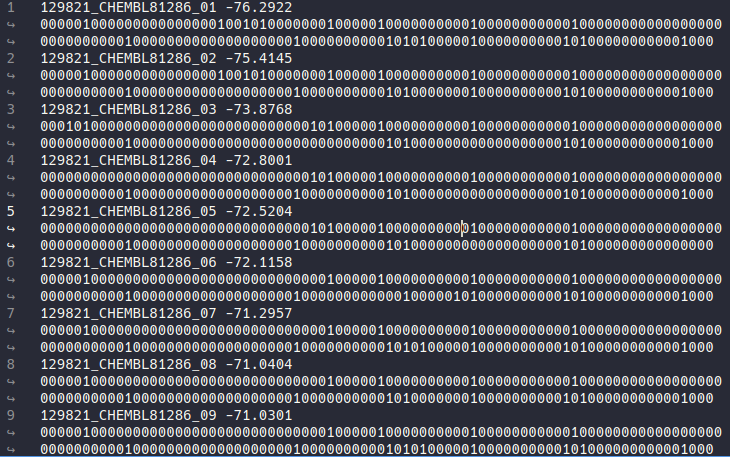
In the next section you will learn how to not only generate the interaction bitstring but also calculate the similarity coefficient using reference bitstring.
Generating Protein-Ligand Interaction Bitstring and Similarity coefficient¶
This procedure will require a reference bitstring which can be generated using hippos-genref included in the package. Open this link to learn how to generate the reference bitstring.
After we acquire the full bitstring we can use it for reference bitstring
as shown in examples/04-na_config_default/config-plants-na-tc-mc.txt which is the configuration file for HIPPOS
docking_method plants # plants or vina
docking_conf ../02-na_plants/plants-003.conf
similarity_coef tanimoto mcconnaughey
full_ref 00000100000000000000000000000000000100000000000001000000000000010000001000000000000000000001000000000000000000000000000000101000000000000000000101000000000010000 00010101000000000000000000000000000100000000000001010000000000010000001000000000000010000000000000000000000001011000001000001000000000000000000101000000000000000 00010101000000100000000000000000000100000000000001010100100000010000001000000000000010000001000000000000010000000000100000101010000000000000000001000000000000000
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
full_outfile plants_full_ifp.csv
sim_outfile plants_similarity.csv
logfile plants.log
Always remember that full_ref should be using the full bitstring from reference. Using bitstring reference of different length will cause an error and the program will stop.
Here the residue_name and residue_number must be the same as the one used for reference
bitstring generation, and you have to set similarity_coef value, such as tanimoto
or mccounaughey or both of them.
docking_method here is plants which correspond to the docking result we would like to
analyse. Then the docking_conf is the configuration file used for docking, HIPPOS require
this file to find the details about docking input and output from PLANTS.
Next, run HIPPOS with the following command inside examples directory:
hippos config-plants-na-tc-mc.txt
there will be 3 output file plants.log, plants_similarity.csv, and plants_full_ifp.csv. The plants_full_ifp.csv will be the same as the one without reference above. The plants_similarity.csv contain the similarity coefficient for every pose comparison. Notice that there are 6 similarity coefficient results which correspond to Tanimoto coefficient and McConnaughey coefficient calculation for 3 reference bitstring
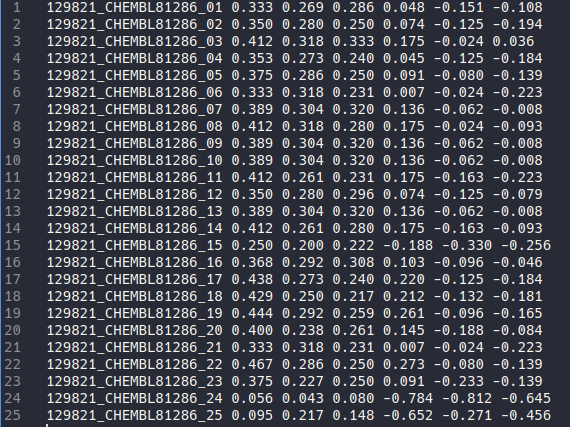
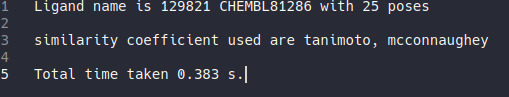
Last but not least the plants.log contain the information about ligand name, number of poses, similarity coefficient used, and the table for bit position for each residue (only appear when output_mode set to simplified, useful for deciphering the simplified bitstring), and the total time taken.
Generating Protein-Ligand Interaction Bitstring and Similarity coefficient (without Backbone)¶
The example above is the default setting where the interaction between ligand and protein backbone
is included. To omit the interaction between ligand and protein backbone, we need to set the
output_mode value to full_nobb, and in order for this setting to work properly we also need to
change the bitstring reference (full_nobb_ref) accordingly
(see how to generate bitstring reference without backbone).
Here is the content of configuration file example examples/05-na_config_nobb/config-plants-na-tc-mc.txt)
docking_method plants # plants or vina
docking_conf ../02-na_plants/plants-003.conf
similarity_coef tanimoto mcconnaughey
output_mode full_nobb
full_nobb_ref 00000100000000000000000000000000000100000000000001000000000000010000001000000000000000000001000000000000000000000000000000101000000000000000000101000000000010000 00010101000000000000000000000000000100000000000001010000000000010000001000000000000010000000000000000000000001011000001000001000000000000000000101000000000000000 00010101000000100000000000000000000100000000000001010000000000010000001000000000000010000001000000000000010000000000100000101010000000000000000001000000000000000
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
full_nobb_outfile plants_nobb_ifp.csv
sim_outfile plants_similarity.csv
logfile plants.log
Always remember that full_nobb_ref should be using the full_nobb bitstring from reference. Using bitstring reference of different length will cause an error and the program will stop.
Like before, run hippos with the following command:
hippos config-plants-na-tc-mc.txt
Just like before, 3 output file will be generated, but the fingerprint (plants_nobb_ifp.csv)
and plants_similarity.csv will be different.
Generating Simplified Interaction Bitstring and Similarity coefficient¶
It is also possible to calculate simplified interaction between ligand and protein. To do so set the
output_mode value to simplified, and in order for this setting to work properly we also need to
change the bitstring reference (simplified_ref) accordingly
(see how to generate simplified bitstring reference).
Here is the content of configuration file example examples/06-na_config_simplified/config-plants-na-tc-mc.txt
docking_method plants # plants or vina
docking_conf ../02-na_plants/plants-003.conf
similarity_coef tanimoto mcconnaughey
output_mode simplified
simplified_ref 0010000000000100000100000110000000010000000000000110000011000000100 0111000000000100000101000110000010000000000111010010000011000000000 0111001000000100000101000110000010010000001000100111000001000000000
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
simplified_outfile plants_simplified_ifp.csv
sim_outfile plants_similarity.csv
logfile plants.log
Always remember that simplified_ref should be using the simplified bitstring from reference. Using bitstring reference of different length will cause an error and the program will stop.
Like before, run hippos with the following command:
hippos config-plants-na-tc-mc.txt
Just like before, 3 output file will be generated, but the fingerprint (plants_simplified_ifp.csv)
and plants_similarity.csv will be different.
Generating Multiple Interaction Bitstring¶
Last but not least, multiple output_mode is also allowed in generation interaction bitstring but without calculation of similarity coefficient. Here is the content of the configuration file example examples/07-na_config_multiple/config-plants-na.txt
docking_method plants # plants or vina
docking_conf ../02-na_plants/plants-003.conf
output_mode full full_nobb simplified
residue_name ARG116 GLU117 LEU132 LYS148 ASP149 ARG150 ARG154 TRP177 SER178 ILE221 ARG223 THR224 GLU226 ALA245 HIS273 GLU275 GLU276 ARG292 ASP294 GLY347 ARG374 TRP408 TYR409
residue_number 40 41 56 72 73 74 78 101 102 145 147 148 150 169 197 199 200 216 218 271 298 332 333
full_outfile plants_full.csv
full_nobb_outfile plants_nobb.csv
simplified_outfile plants_simplified_ifp.csv
logfile plants.log
Like before, run hippos with the following command:
hippos config-plants-na.txt
Now, four output file will be generated, three for three different output, and one for the log file.